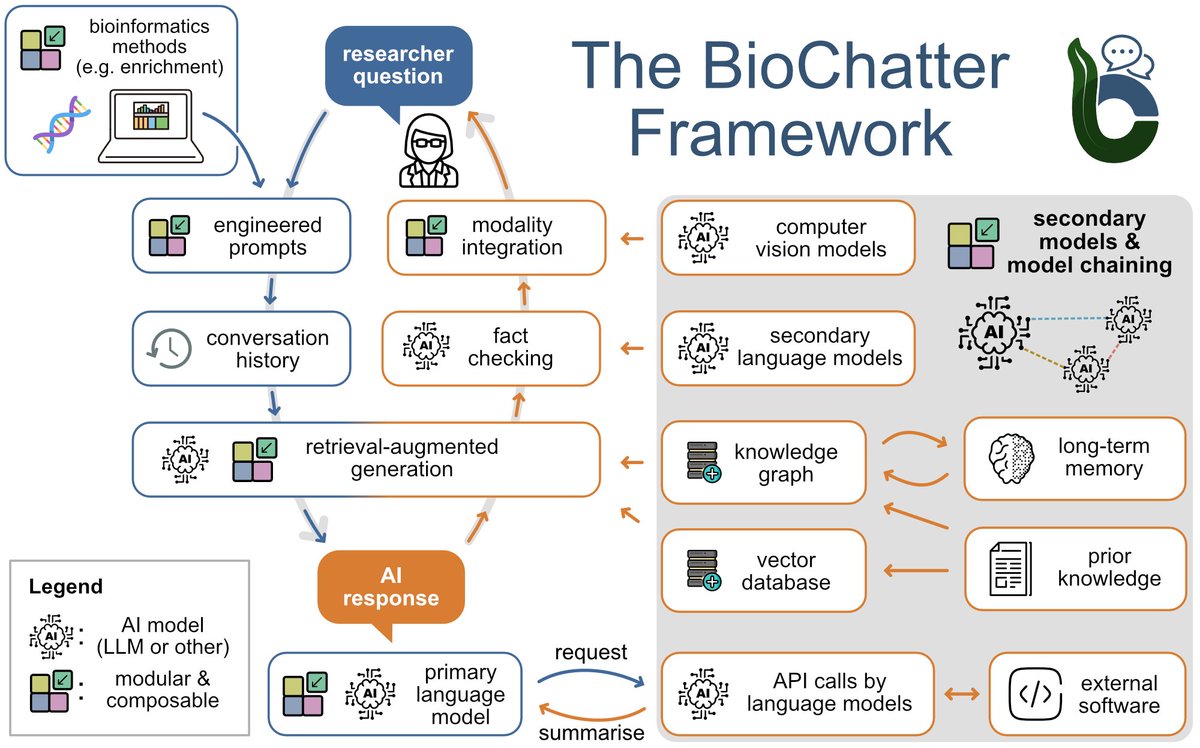
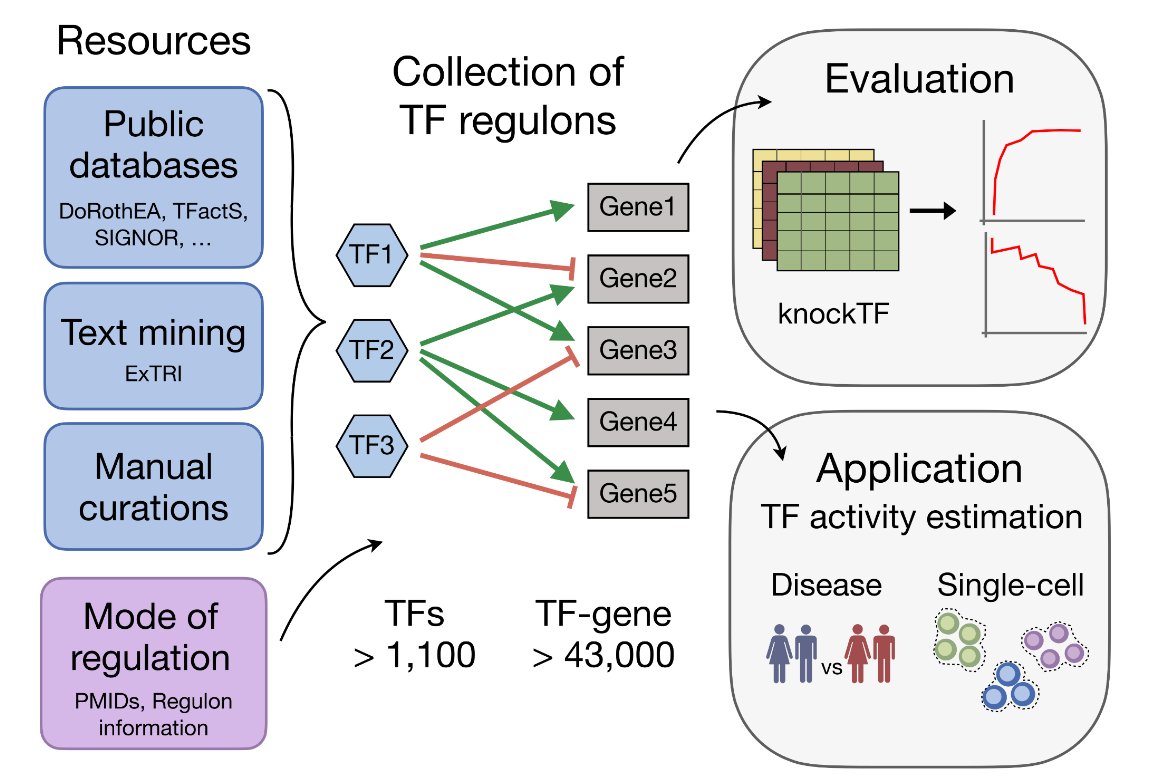
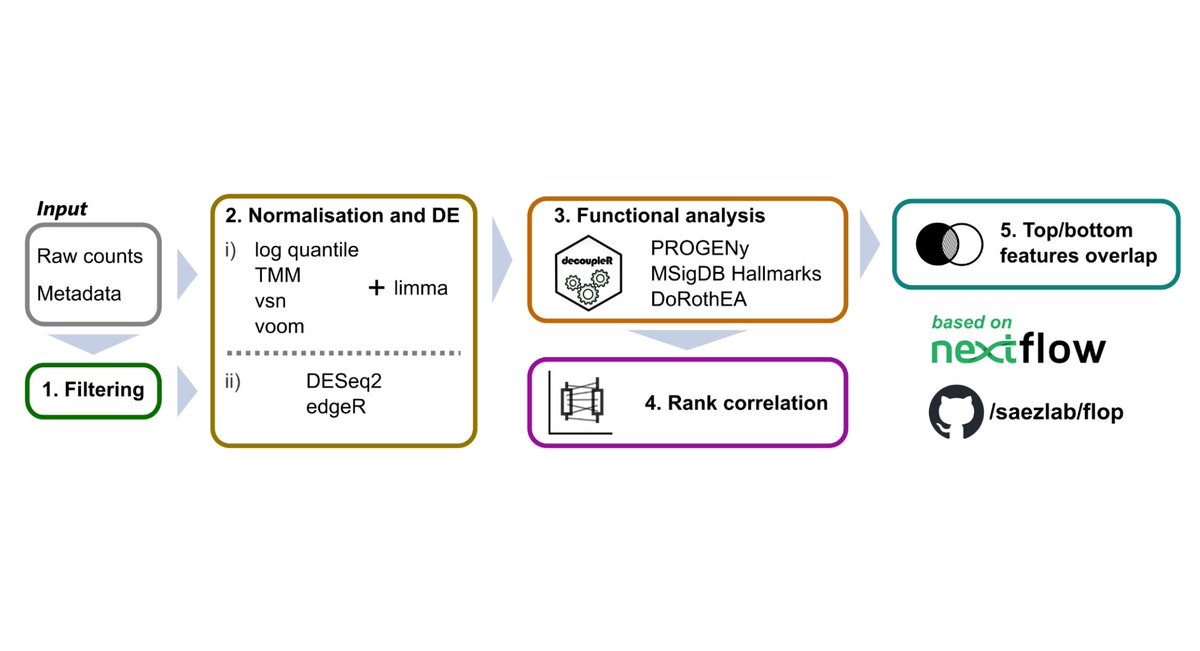
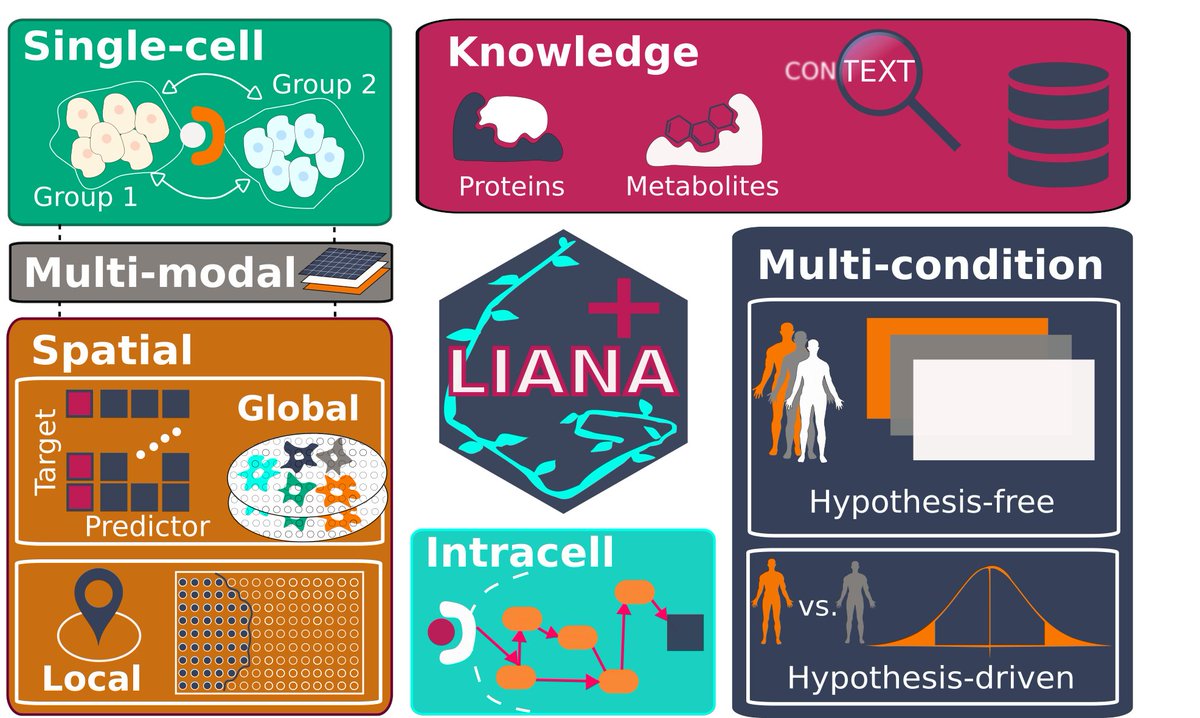
Julio Saez-Rodriguez
@JulioSaezRod
Computational Biologist, Head of research @emblebi, part-time @UniHeidelberg, codirector @DR_E_A_M challenges. Low-activity account, mostly retweets @saezlab
ID:264914367
http://saezlab.org 12-03-2011 17:29:55
260 Tweets
2,2K Followers
381 Following



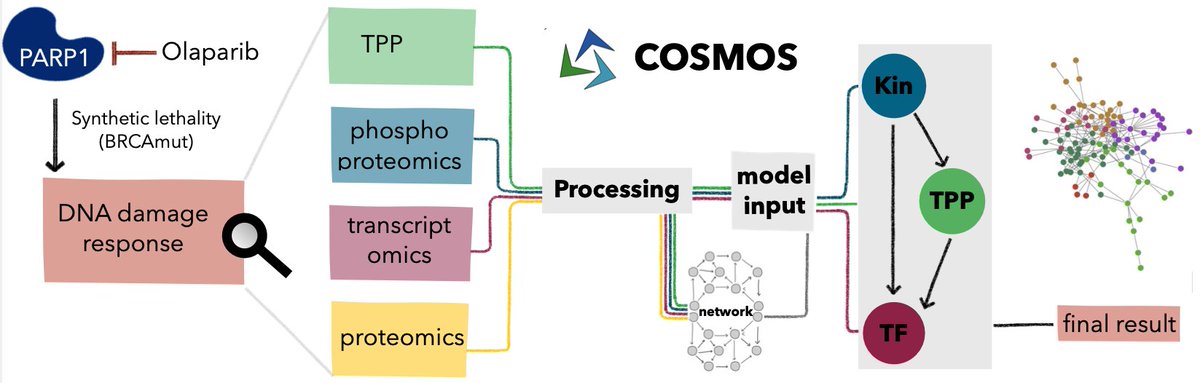
Our study integrating functional proteomics, phosphoproteomics and transcriptomics using COSMOS is out in Mol Syst Biol 🧬🧫
You can find it here: tinyurl.com/COSMOS-TPP
More information in the thread below 👇


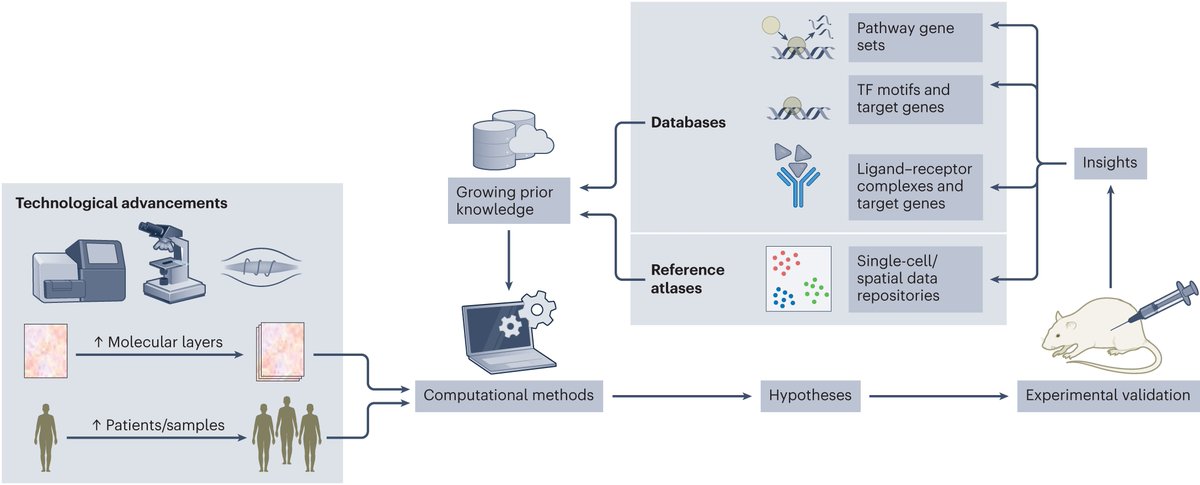
1) Are you interested in computational approaches to study the immune system 🔎🦠by integrating single-cell + spatial multi-omics data with prior knowledge? Check our review 📖out now in Nature Immunology nature.com/articles/s4159… 🧵


Register to listen March 7th to our Julio Saez-Rodriguez talk about 'large-scale models of signaling from prior knowledge and omics: where we are and where we may go to', along w Ruth Huttenhain 's talk on spatially resolved signaling

If you do image analysis, check these recommendations & pitfalls - Congrats Lena Maier-Hein Annika Reinke Paul F. Jäger Minu Dietlinde Tizabi + rest of the team and thanks for involving our Julio Saez-Rodriguez

Our Julio Saez-Rodriguez will give #biomedx #lunchtalk Thursday, January 25, 11 AM Communication Center DKFZ #Heidelberg about 'Knowledge-Based Machine Learning to Extract Disease Mechanisms from Omics Data' bio.mx/news/events/

Congrats Markus Rinschen and team and thanks for involving us! Glad our Aurelien Dugourd R. Fallegger Julio Saez-Rodriguez could help to analyse this multi-omics data to understand the effect of SGLT2 inhibitors

#spatial technologies advance v rapidly - See 👇 Mol Syst Biol from Theodore Alexandrov Sinem Saka & our Julio Saez-Rodriguez review & perspective on major challenges & opportunities, incl. computationally

Congrats Rebekka Schneider Hélène Gleitz Stephani Schmitz and thanks for involving our Aurelien Dugourd Julio Saez-Rodriguez in the analysis of this very interesting data.


Thanks to funding from DFG public | @[email protected] 🙏 we will soon be recruiting developers to extend our prior knowledge frameworks BioCypher (biocypher.org) and OmniPath (omnipathdb.org): ssc.uni-heidelberg.de/en/newsroom/tw….

first European Research Council (ERC) funding after 7 applications and 5 interviews❗ so, if you did not get it, don’t give up! And, if you can, get as awesome partners as Stefania Giacomello Eduardo Villablanca Zeller Group 😄 Very thankful to them, many Saez-Rodriguez Group members & all others that helped us🙏




Different omics provide complementarity information - we explored how to integrate them to understand PARP inhibition in ovarian cancer using our multi-omics network framework COSMOS in our latest work led by Mira Burtscher (tinyurl.com/579fehuy)

We’ll be at #ISMBECCB2023 !👇
Ece Kartal Predicting HF from microbiome🫀🦠
Pau Badia i Mompel Evaluation of gene regulatory nets🧬
Julio Saez-Rodriguez Knowledge-based machine learning🖥️
Loan Vulliard Tumor metabolic environment imaging📸
@TimTheRose MS annotation using metabolic nets✍️