Computational Biology, Group Leader @goetheuni Frankfurt
@InstCardReg_FFM
ID:717569454503231488
https://schulzlab.github.io 06-04-2016 04:27:44
4,2K Tweets
978 Followers
476 Following

A study in Nature Microbiology shows that enzymes found in the gut resident bacteria, Akkermansia muciniphila, can convert both known and previously not recognized antigens on human red blood cells to make group O blood. go.nature.com/3QLIPTh



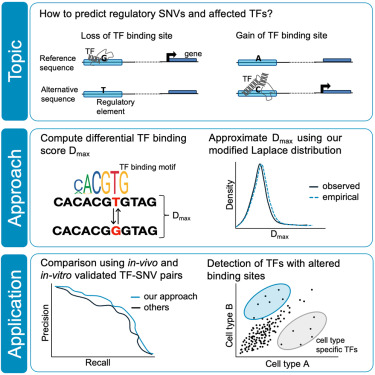
A statistical approach for identifying single nucleotide variants that affect transcription factor binding Marcel Schulz - @[email protected]
cell.com/iscience/fullt…

Today first seminar of the critical #genomics initiative at Goethe-Universität . Prof. Dorothea Schulte talked about #DNA and how it decodes organisms. Every week on Wednesday a new speaker will address another topic including Anna Y. Yotova . More info here: critical-genomics.de/index_de.html






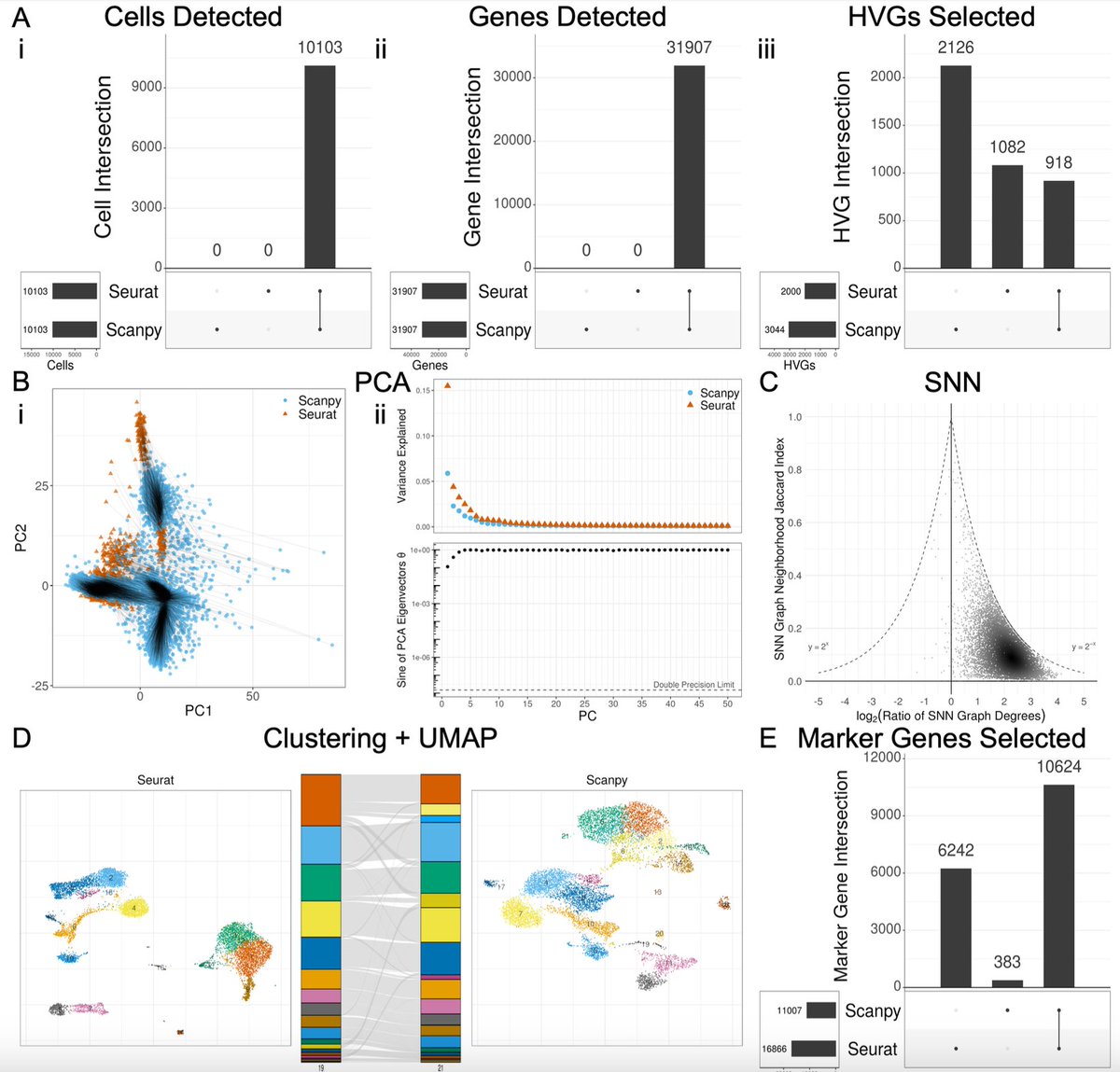
As my lab develops Seurat, I wanted to share some thoughts on the important Seurat and scanpy package selection preprint from Joseph Rich/Lior Pachter 🧵

The choice of whether to use Seurat or Scanpy for single-cell RNA-seq analysis typically comes down to a preference of R vs. Python. But do they produce the same results? In biorxiv.org/content/10.110… w/ Joseph Rich et al. we take a close look. The results are 👀 1/🧵

what a birthday present! 💃🏻💃🏻 Our paper characterising pericytes in cardiac ageing has just been published by Circulation Research -> ahajournals.org/doi/10.1161/CI… Special kudos to future Dr. Anita Tamiato who did the heavy lifting 💪🏻💪🏻





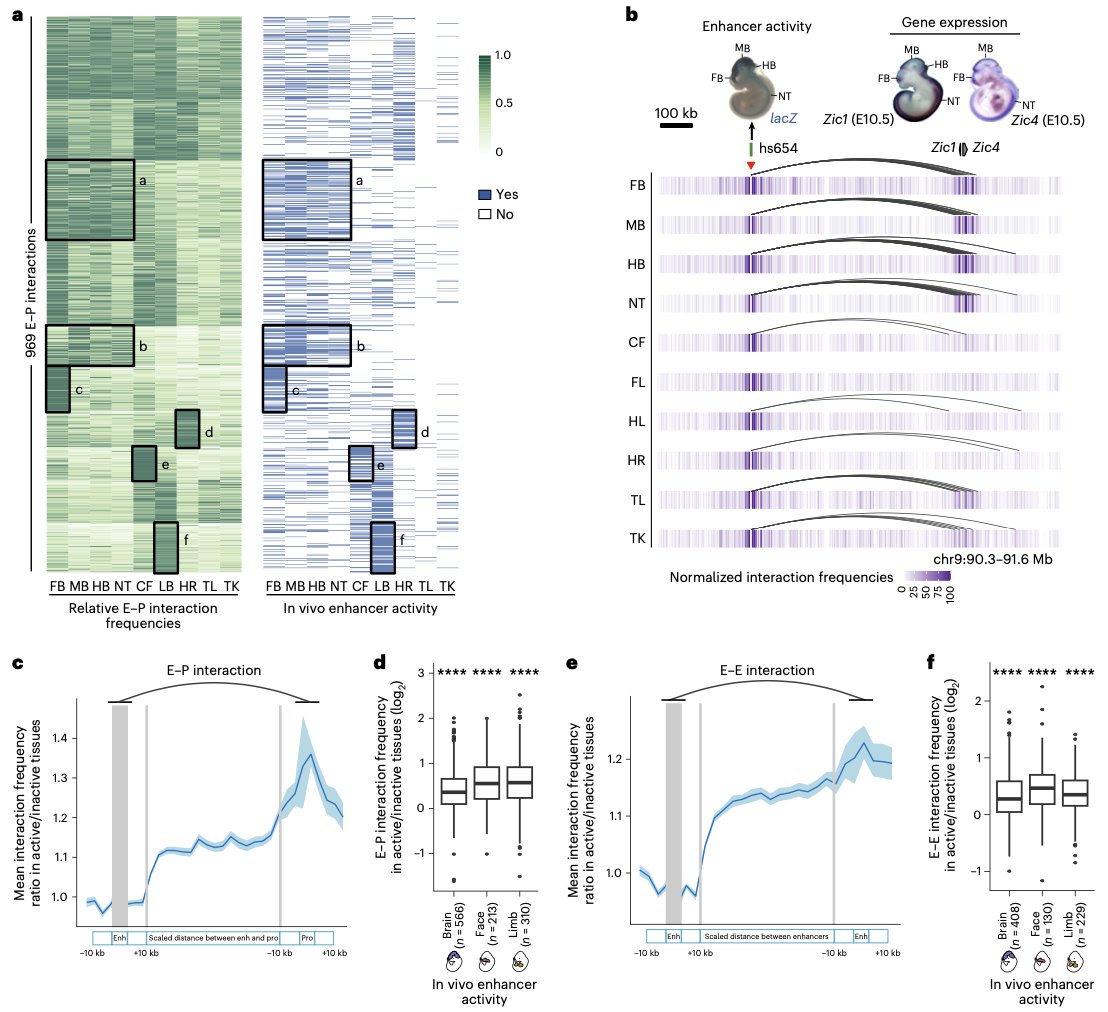
Our preprint characterizing mouse developmental Enhancer—Promoter interactions by Zhuoxin is out today Nature Genetics rdcu.be/dBRPu
See also a complementary study on fly Enhancer—Promoter interactions from Eileen Furlong nature.com/articles/s4158…

NOW LIVE ON TAPE
Our Human Cell Atlas Webinar
“Focus on Technology”
Featuring: Spatial genomics, Lineage tracing, Perturbation and AI tools.
Starring: Samantha Morris, Nikolaus Rajewsky, Maria Brbic, Eric Lubeck.
Enjoy 👇👇👇 youtube.com/watch?v=fI9Oe_…

Our most recent work is now published at Nature Genetics. Epigenetic variation impacts individual differences in the transcriptional response to influenza infection. Great collab with Guillaume Bourque lab and huge congrats to the lead author Katie Aracena. Link: shorturl.at/bcdBP