The DECIPHER Project
@deciphergenomic
Enabling #matchmaking and interpretation of phenotype-linked pathogenic genetic variation in patients with #raredisease by effective #datasharing.
ID:4501798349
https://www.deciphergenomics.org/ 16-12-2015 10:11:03
609 Tweets
2,0K Followers
108 Following


.The DECIPHER Project v11.25 is here.
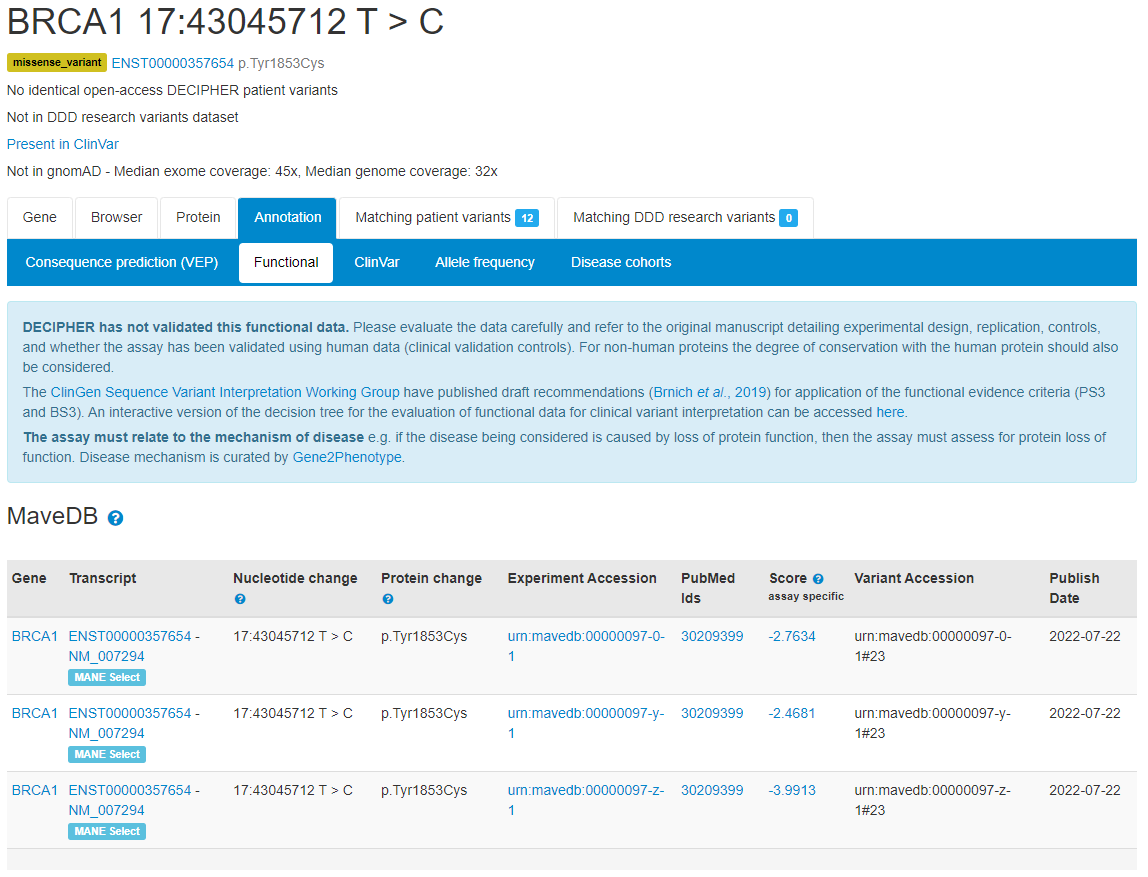
This update introduces new features including the display of functional data from multiplexed assays of variant effects (MAVEs) to assist variant interpretation and much more.
ebi.ac.uk/about/news/upd…


Dom McMullan 💙 Matt Hurles The DECIPHER Project We LOVE Decipher!!!! My total go-to for preliminary looking at variants. Wish I had more time to make better use of this brilliant and intuitive tool.





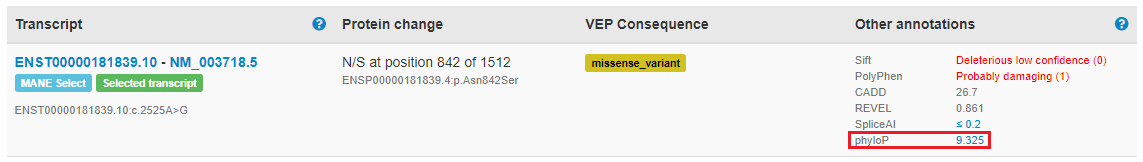
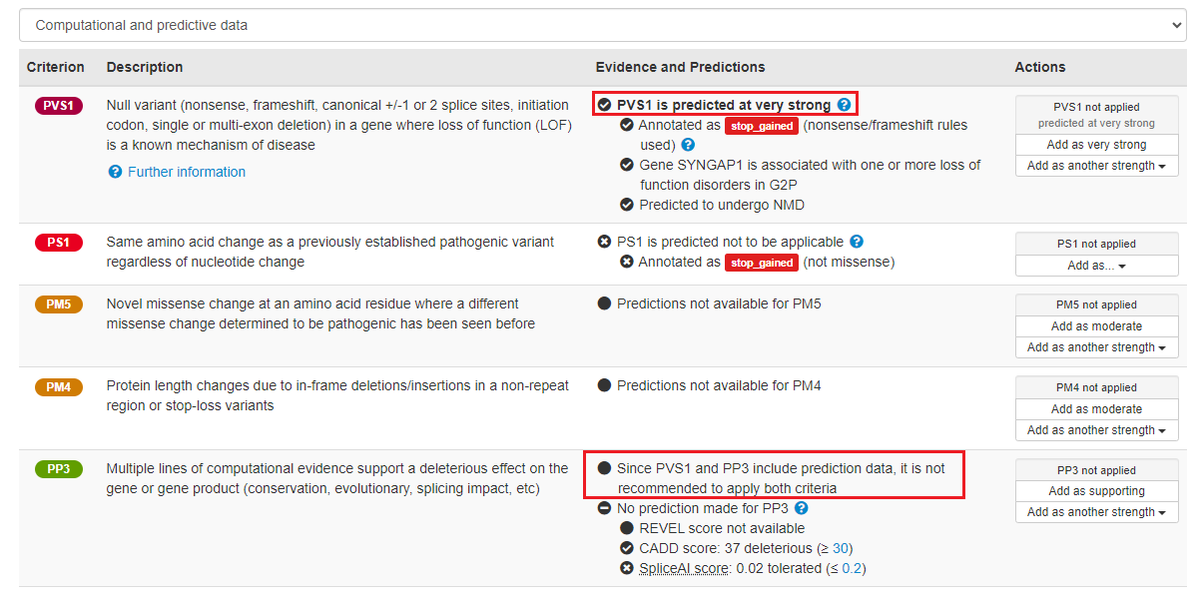
A link to the recommendations for clinical interpretation of variants found in non-coding regions of the genome is displayed in the sequence variant pathogenicity interface when a variant in a non-coding region is being evaluated Jamie Ellingford Nicky Whiffin #noncoding



Functional data from Multiplexed Assays of Variant Effect (MAVEs) from the MaveDB repository are now displayed on functional tabs. Links to experimental design and score sets in MaveDB are provided, in addition to publication(s) links Atlas of Variant Effects Alliance Alan Rubin

DECIPHER version 11.25 has been released. See the new features at deciphergenomics.org #variantinterpretation


Last year, The DECIPHER Project made the move over to EMBL-EBI after many successful years at Wellcome Sanger Institute and this year DECIPHER celebrates its 20th anniversary 🎉
Find out more about DECIPHER and the move over to EMBL-EBI.
ebi.ac.uk/about/news/ann…

This #RareDiseaseDay we’re shining the spotlight on The DECIPHER Project, our database for genetic and phenotypic data which helps further our understanding of rare diseases and improving patient diagnoses.
Find out more 👇
deciphergenomics.org


Thank you Ryan Collins for generating and sharing pHaplo and pTriplo scores - now displayed as tracks in the UCSC Genome Browser