Marinka Zitnik
@marinkazitnik
Assistant Professor at Harvard | Faculty @Harvard @KempnerInst AI | Faculty @broadinstitute @harvard_data | Cofounder @ProjectTDC | @AI_for_Science
ID:61502457
https://zitniklab.hms.harvard.edu 30-07-2009 14:34:38
2,7K Tweets
6,3K Followers
226 Following




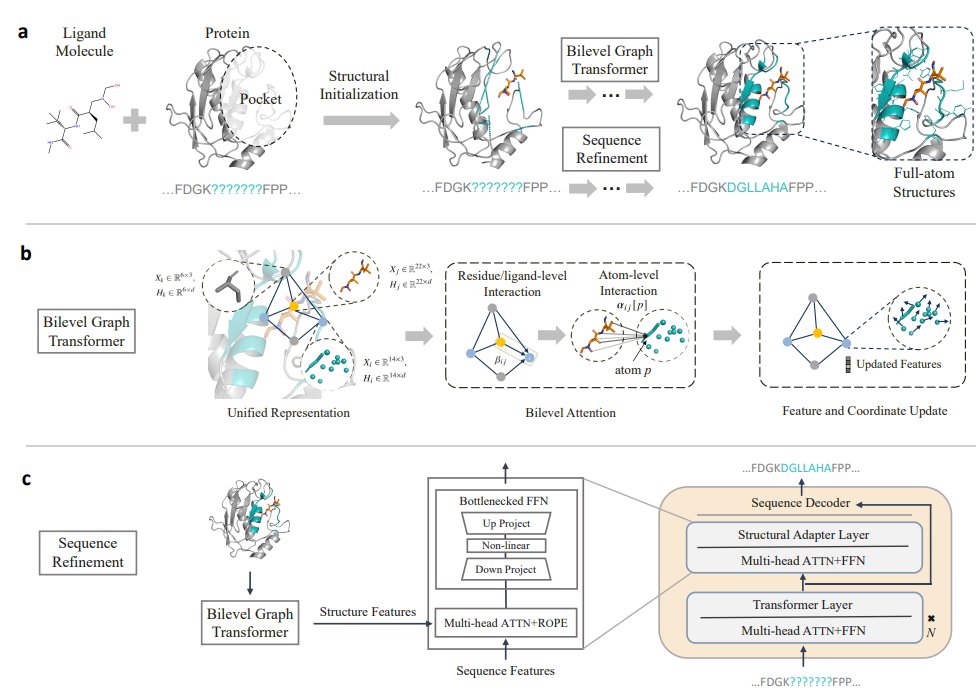
The release of AlphaFold 3 by Google DeepMind and Isomorphic Labs is a big leap in the prediction of a wider universe of biomolecules.
But, its usefulness for drug discovery, and impact on trial failure rates, is a big open question.
statnews.com/2024/05/08/dru… via STAT

Join me on Sat 12-2pm at the MLGenX Workshop @ ICLR 2024 poster session to talk about PDGrapher, our work on predicting therapeutic perturbations:
openreview.net/forum?id=Zcw2O…
I’m at #ICLR2024 Thurs to Sat - reach out to talk (graph) ML + Bio or about our Prescient Design team!


🦾 AI for Science: Scaling in AI for Scientific Discovery, co-organized by HDSI Faculty Affiliate Marinka Zitnik, will feature speakers recognized for their understanding of scaling’s impact on AI for Science: ai4sciencecommunity.github.io/icml24.html
⏳ Abstract Submission Deadline: May 23, 2024


That's a wrap on this semester's DBMI at Harvard Med BMI702 Intro to #Biomedical #AI class! 🎓
Explore the syllabus and course materials, open to all: zitniklab.hms.harvard.edu/BMI702/ Harvard Medical School Harvard Data Science Initiative Kempner Institute at Harvard University