First author JCO pub. We identify that germline mutations in ATM,BRCA2,CDH1,CHEK2 & PALB2 are associated with increased risk of invasive lobular carcinoma of breast, whereas BRCA1 is not.
Mayo Clinic Comprehensive Cancer Center Mayo Clinic Hematology-Oncology Fellowship BasserBRCA ASCO Publications OncoAlert
ascopubs.org/doi/abs/10.120…

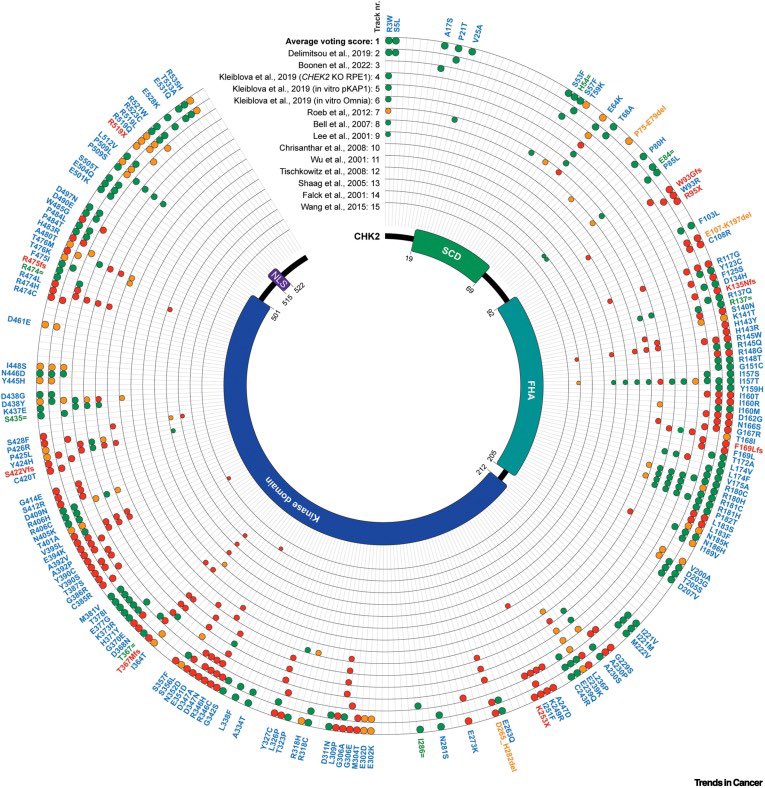
What can #CHEK2 teach us about moderate risk? The mutation matters. Mark Robson City of Hope #cohgenomics #gcchat #CancerGenetics


#TumorBoardTuesday Mike Pishvaian Shilpa Gupta Andrey Soares Martín Angel Misha Beltran Edwin Posadas, MD AttardLab Chris Parker Dimitrios Giannis MD, MSc Eleni Efstathiou Matt Sydes Alicia Morgans, MD, MPH Josh Lang Jason Brown Prateek Mendiratta JAGarciaMD Charbel Hobeika, MD Karine Tawagi MD Gianluca Giannarini Simon C Tom Varghese Jr. MD, MS, MBA, FACS, MAMSE 🇺🇸 Johnathan Ebben MD, PhD @gulleyj1 Uromigos Colin Pritchard Petros Grivas Ian Davis Arun Azad Andrea Miyahira Benjamin L Maughan Dan George Alan H Bryce Andy Hahn 𝐂𝐡𝐚𝐧𝐝𝐥𝐞𝐫 𝐏𝐚𝐫𝐤 𝐌𝐃 𝐅𝐀𝐂𝐏 Nazli Dizman Abhenil Mittal Cristiane D Bergerot Atish Choudhury, MD PhD Minas Economides, MD Jonathan Rosenberg MD Abhishek Tripathi Brian Rini, MD Yüksel Ürün Hans Hammers 4/18 #TumorBoardTuesday Amanda Nizam, MD Vinay Mathew Thomas Alejandro Sanchez, MD Joshua Meeks
#ProstateCancer
👩🏽🏫Mini tweetorial 2👨🏻🏫
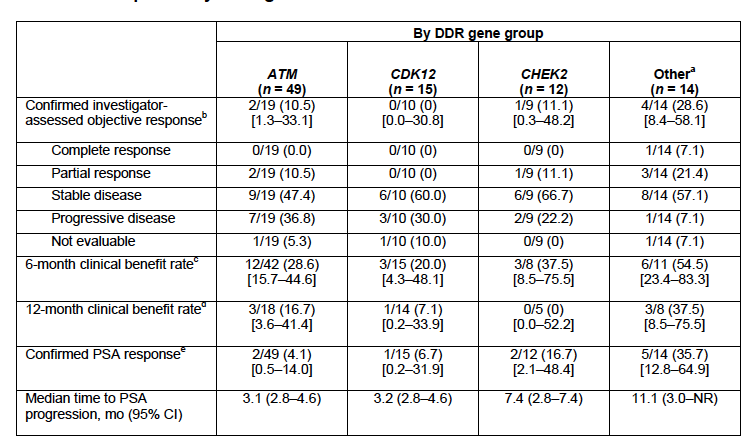
📌mCRPC patients with DNA repair alterations
📍Majority were HRR mutations
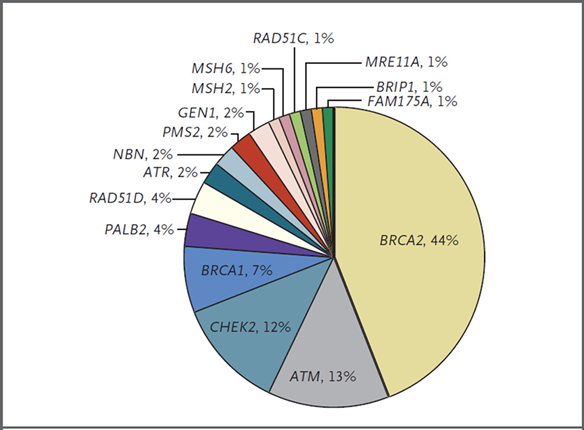
📍BRCA2 44%
📍ATM 13%
📍CHEK2 12%
📍BRCA1 7%
📚Joaquin Mateo ncbi.nlm.nih.gov/entrez/eutils/…

23% of all #patients w/ #testicularcancer #germlinetesting had pathogenic mutations, 19% in DNA-damage repair genes (e.g. ATM, CHEK2). #DrChethanRamamurthy UT Health San Antonio Guru P. Sonpavde, MD Dana-Farber Invitae Urologic Oncology
#access #genetictesting #clinicaltrials
authors.elsevier.com/c/1eOZ23r93nRQ…
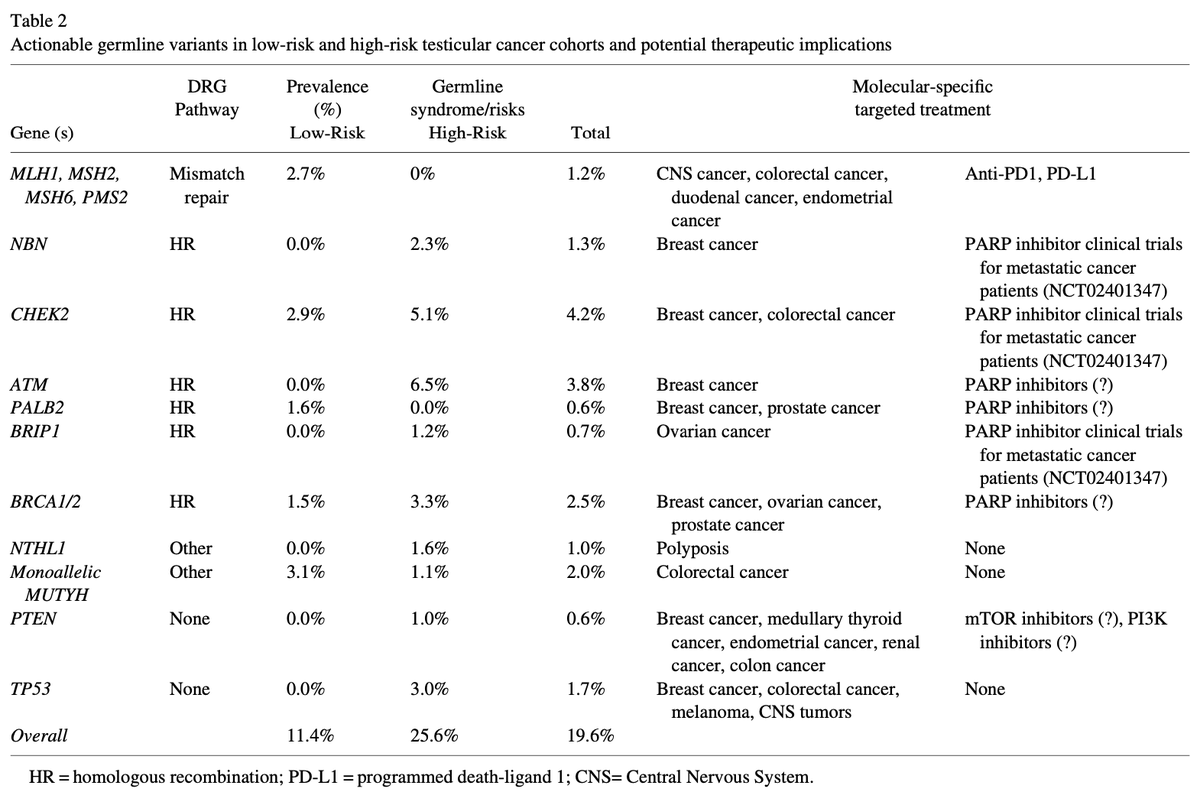

⚡️Can you give PARP inhibitor alone (WITHOUT ADT) for BRCA1, BRCA2, or CHEK2 metastatic hormone sensitive prostate cancer patients?
ORR 60% for BRCA2 patients. Small study but very intriguing by #MarkMarkowski and #EmmanuelAntonarkis
ASCO #GU23 #PosterSession
OncoAlert




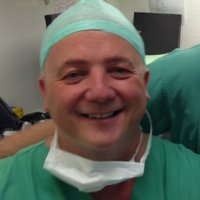



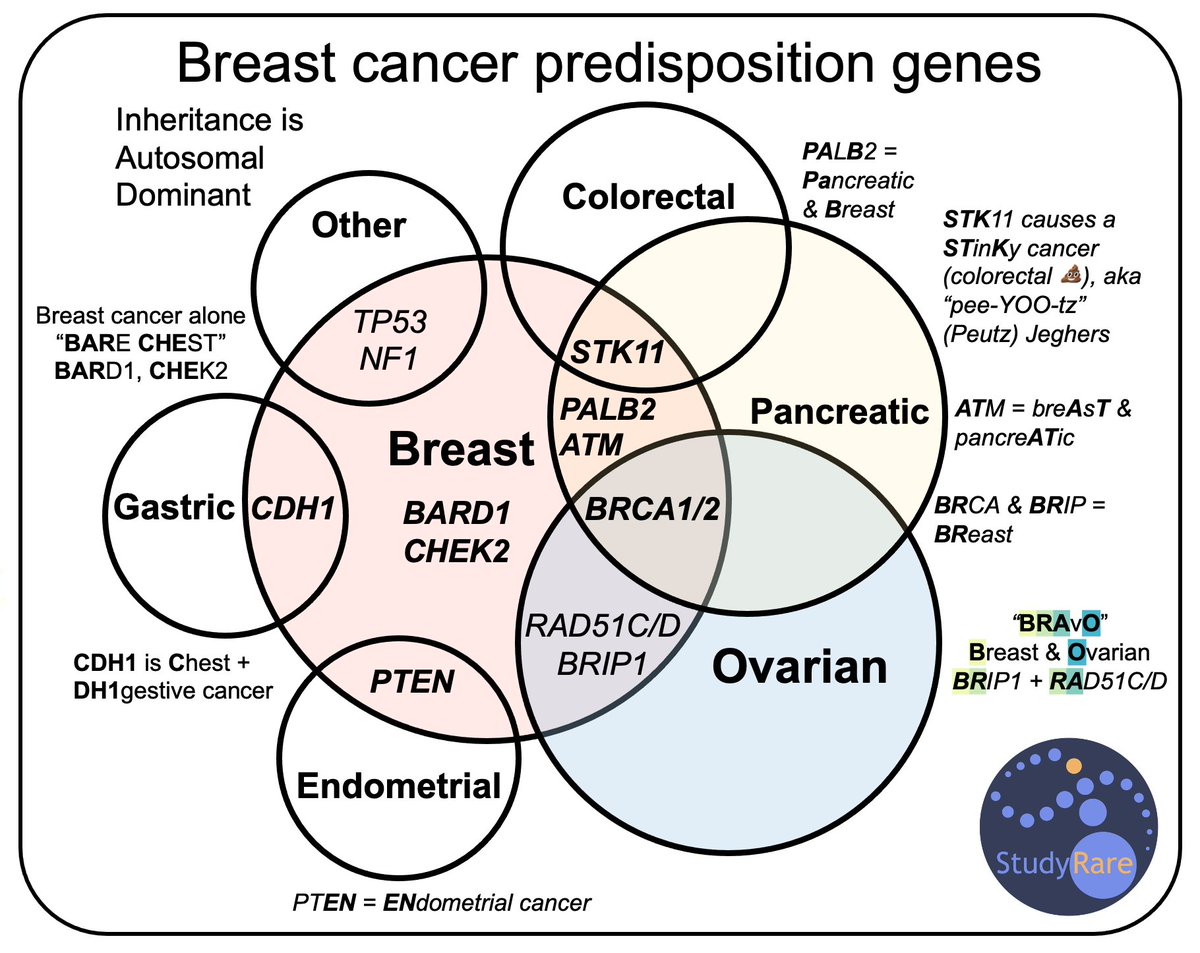
Interestingly, women carrying germline pathogenic variants in ATM, BRCA, BRCA2, CHEK2, and PALB2 have an increased risk of #breastcancer and may benefit from improved surveillance and risk reduction strategies.
ascopubs.org/doi/full/10.12…




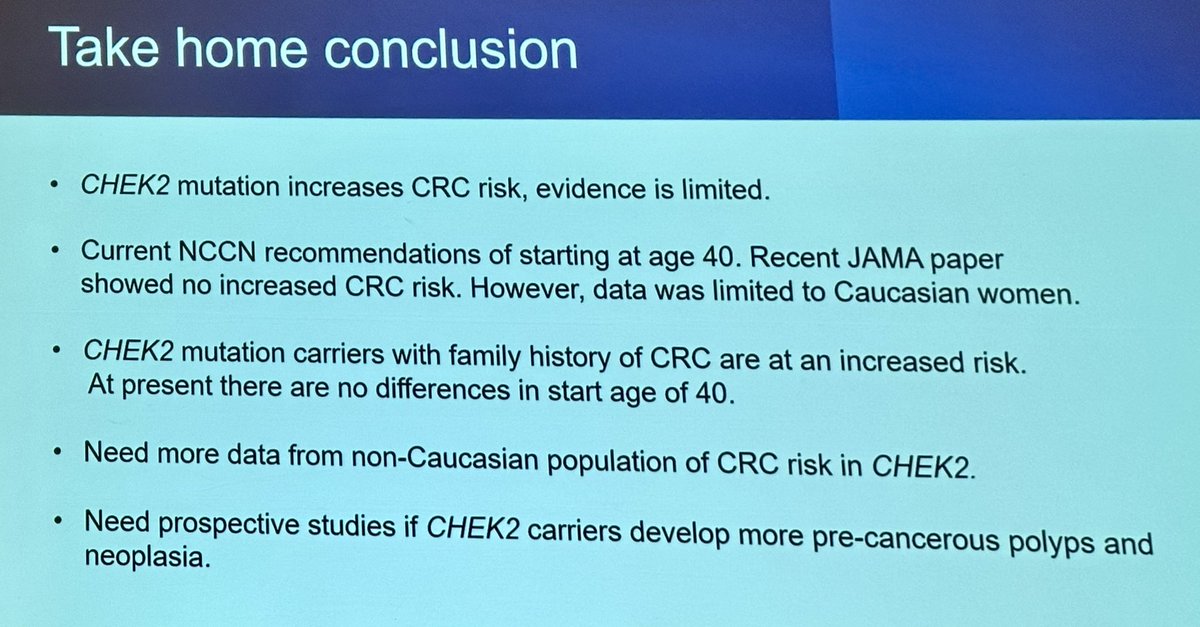
👉🏻Still more questions than answers in the role of CHEK2 in #CRC predisposition
➡️Until more answers comes…the recommendation is CRC screening as for high-risk familial CRC National Comprehensive Cancer Network (NCCN)
👏🏻👏🏻Nice presentation by Priyanka Kanth,MD,MS CGA-IGC #CGAIGC22