I'm excited to share the peer-reviewed version of my PhD work published today in Cell
Check out our video where we explain the main findings of our research. 1/3
#BlastOFF #Proteomics #ScienceTwitter
cell.com/cell/fulltext/…

Super excited to get my first set of diatom spatial proteomics samples on the mass specs Woods Hole Oceanographic Institution (WHOI). One part of the sample on the Orbitrap Astral for proteomics and another part on the iCAP for trace metal measurements!!


A recent top Pastel BioScience #prot eomics post, as provided by Twitter analytics …
x.com/pastelbio/stat…
---
#prot eomics #prot -other


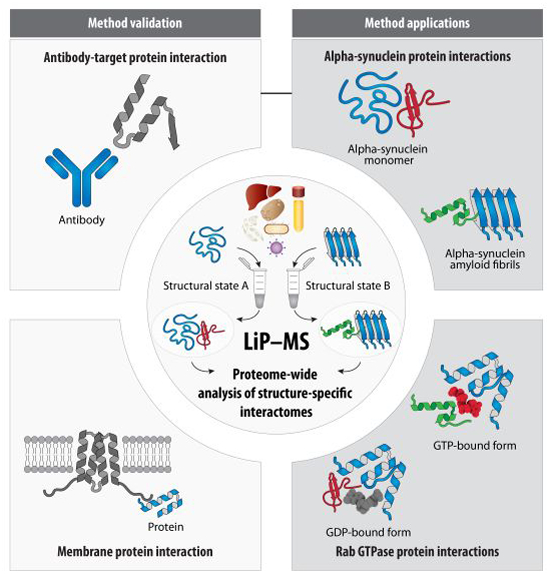
Our paper describing a method to screen for conformation-specific protein-protein interactions is out Mol Syst Biol.
Read it here: doi.org/10.1038/s44320…
The approach can also be used to map interaction sites.
IMSB_ETH ETH Zürich ETH Zürich_en
#proteomics #massspectrometry #interactomes










A great team effort led by Neftaly Cruz Mireles & proteomics expert Frank Menke & including Miriam Oses-Ruiz Lauren Ryder Bozeng Tang EseolaAlice Jave Bautista Were Vincent Paul Derbyshire, Clara Jegousse, Dan Maclean & Kim Findlay


Great visit with Peter Black learning about new clinical trial design in muscle invasive bladder cancer based on subtypes, and role of proteomics and nothing beats a morning bicycle ride in scottsdale Joshua Meeks Seth lerner Petros Grivas Mark Tyson


A recent top Pastel BioScience #prot eomics post, as provided by Twitter analytics …
x.com/pastelbio/stat…
---
#prot eomics #prot -other