Winston Timp
@timp0

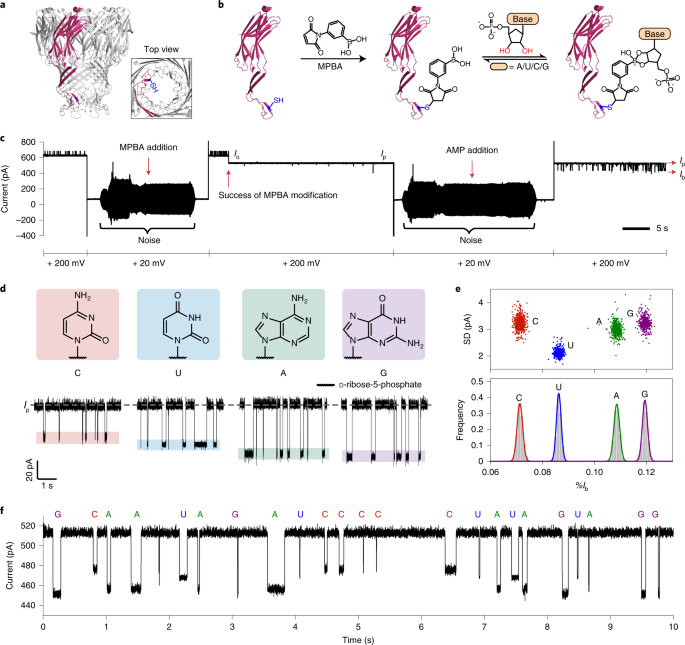
Our preprint is out! We hacked the Oxford Nanopore sequencer to read amino acids and PTMs along protein strands. This opens up the possibility for barcode sequencing at the protein level for highly multiplexed assays, PTM monitoring, and protein identification!
biorxiv.org/content/10.110…

What do you do after a negative exome? So happy to see this paper out which covers some of the new technologies we're using in GREGoR Consortium to evaluate challenging unsolved cases! sciencedirect.com/science/articl…

Beyond assembly: the increasing flexibility of single-molecule sequencing technology go.nature.com/3HSiBd4 #Review by Paul W. Hook & Winston Timp Johns Hopkins University Johns Hopkins BME

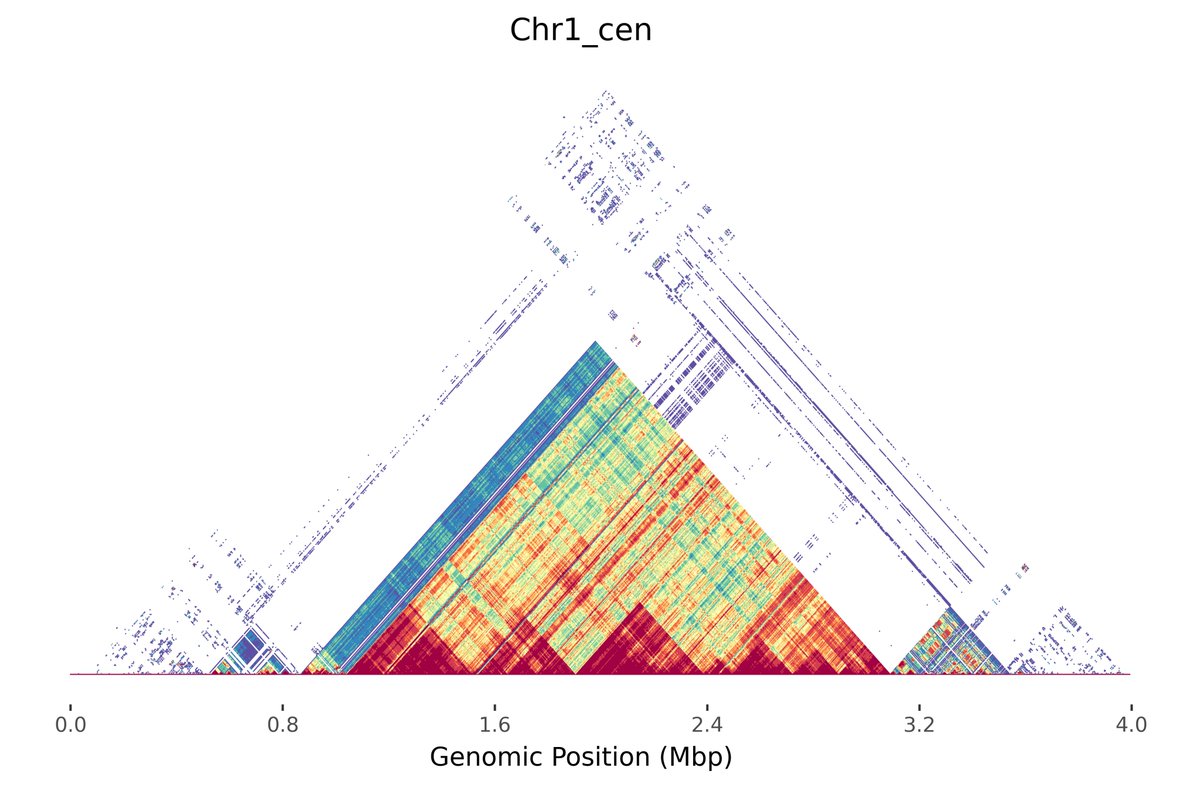
Check out the beta release of Alex Sweeten's new tool Mod.Plot! We 'mashed up' Mitchell R. Vollger's StainedGlass visualization with minhashing to enable instant and interactive dotplotting. No alignment needed! Preprint of the method to follow in a couple months 📈



So thrilled the Lustgarten Foundation is supporting this cutting-edge project to unlock the inherited genetic basis of pancreatic cancer using long-read sequencing. My co-leaders Michael Schatz Winston Timp are true pioneers in this area. WE ARE HIRING!

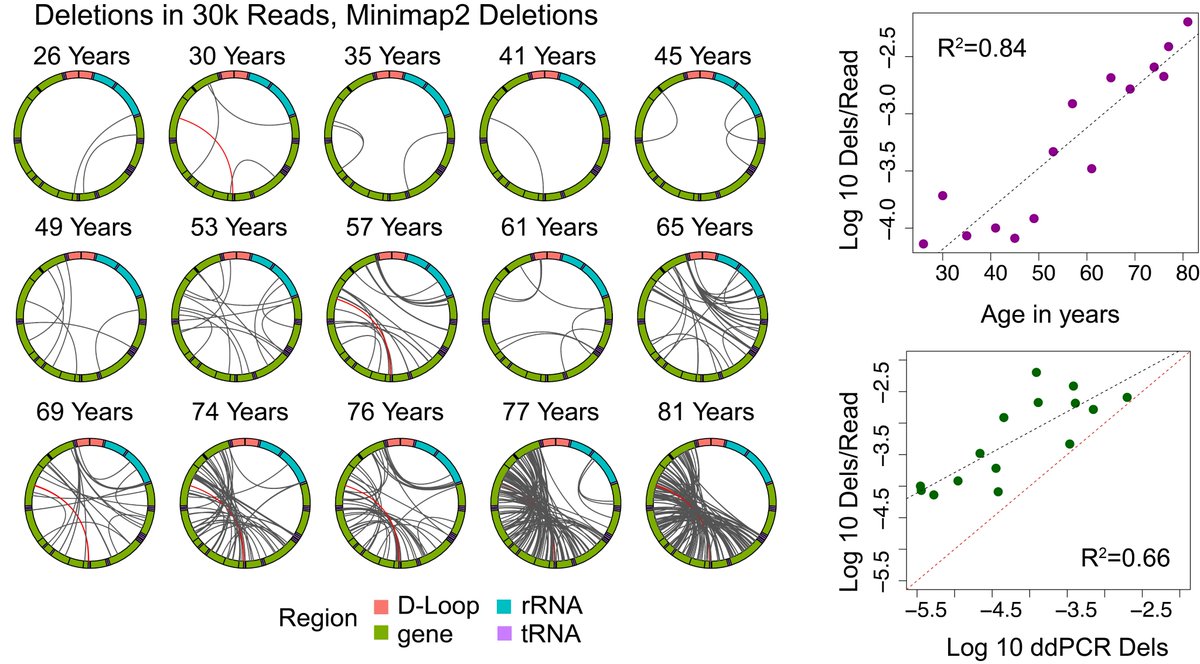
These data provide invaluable information for future work to understand the mechanisms underlying formation and amplification of mitochondrial genome deletions and for using these variants as a metric of aging. Thank you to Winston Timp for collaboration!



We at PacBio are finally presenting to the world, at long last, isoform information at #singlecell resolution - some details on the tech background for the kit and #Bioinformatics below 🧵

Congratulations Justus, Johns Hopkins University recipient of a 2022 Klingenstein-Simons Fellowship Award in Neuroscience for the project “Mechanisms of evolutionary circuit expansion,” bit.ly/3LMrSUH Simons Foundation



How quickly can Oxford Nanopore be used to assess genetic risk? In this preprint we show that we could determine whether a newborn inherited two known familial variants within 3 hours of birth by WGS and targeted analysis 1/n:
medrxiv.org/content/10.110…

Long-read scRNA-seq is awesome for studying RNA isoforms. But Oxford Nanopore has relied on matched short-reads to identify cell barcodes and assign reads to cells.
Pleased to share BLAZE, a new tool to identify cell barcodes from nanopore scRNA-seq. Short-reads not required. 1/4

Excited to share our paper, out today in nature, where we characterize transcriptome variation in human tissues by long-read sequencing. Huge thanks to Dafni Beryl Cummings Garrett Garborcauskas Daniel MacArthur and other collaborators. 🧵
nature.com/articles/s4158…

Genome editing + nanopore sequencing (DNA, RNA, and yes even protein sequencing too!) Oh my! Apply here to work with Thomas Gorochowski and I on an exciting new synbio venture👇🏼


Happy to share our latest work from Winston Timp lab, modbamtools: Analysis of single-molecule epigenetic data for long-range profiling, heterogeneity, and clustering
👉manual and tutorial:
rrazaghi.github.io/modbamtools/
👉preprint: biorxiv.org/content/10.110…
some of the tools to highlight: 1/

I am so thankful to have such an awesome lab, such amazing colleagues, phenomenal support staff, and incredibly supportive mentors, friends, and family! Cheers Michael! QB3-Berkeley CCB UC Berkeley